Accessing all profiles at once#
Conveniently, all the core mission profiles are compacted in a single file, named: <FloatWmoID>_prof.nc. However, some information is only in the individual profile files, but we will see it.
Let’s start loading the neccesary python modules and the netCDF file 6901254/6901254_prof.nc
import numpy as np
import xarray as xr
import netCDF4
from matplotlib import pyplot as plt
%matplotlib inline
prof = xr.open_dataset('../../Data/202107-ArgoData/dac/coriolis/6901254/6901254_prof.nc')
prof
<xarray.Dataset> Size: 892kB
Dimensions: (N_PROF: 104, N_PARAM: 3, N_LEVELS: 98,
N_CALIB: 1, N_HISTORY: 0)
Dimensions without coordinates: N_PROF, N_PARAM, N_LEVELS, N_CALIB, N_HISTORY
Data variables: (12/64)
DATA_TYPE object 8B ...
FORMAT_VERSION object 8B ...
HANDBOOK_VERSION object 8B ...
REFERENCE_DATE_TIME object 8B ...
DATE_CREATION object 8B ...
DATE_UPDATE object 8B ...
... ...
HISTORY_ACTION (N_HISTORY, N_PROF) object 0B ...
HISTORY_PARAMETER (N_HISTORY, N_PROF) object 0B ...
HISTORY_START_PRES (N_HISTORY, N_PROF) float32 0B ...
HISTORY_STOP_PRES (N_HISTORY, N_PROF) float32 0B ...
HISTORY_PREVIOUS_VALUE (N_HISTORY, N_PROF) float32 0B ...
HISTORY_QCTEST (N_HISTORY, N_PROF) object 0B ...
Attributes:
title: Argo float vertical profile
institution: FR GDAC
source: Argo float
history: 2021-07-09T12:28:17Z creation
references: http://www.argodatamgt.org/Documentation
user_manual_version: 3.1
Conventions: Argo-3.1 CF-1.6
featureType: trajectoryProfileIn this case, N_PROF is 104, since there are 104 profiles, including two for the first cycle, the descending and the ascending. The profiles in _prof.nc are just the ‘Primary sampling’.
If you need the high-resolution upper 5 dbar you have to use the individual cycle files, LOADING ‘../../Data/202107-ArgoData/dac/coriolis/6901254/profiles/R6901254_001.nc’ as we saw in the previous chapter.
Let’s visualize all the salinity observations, a first a quick-look
fig , ax = plt.subplots(figsize=(10,10))
ax.contourf(prof.PSAL);
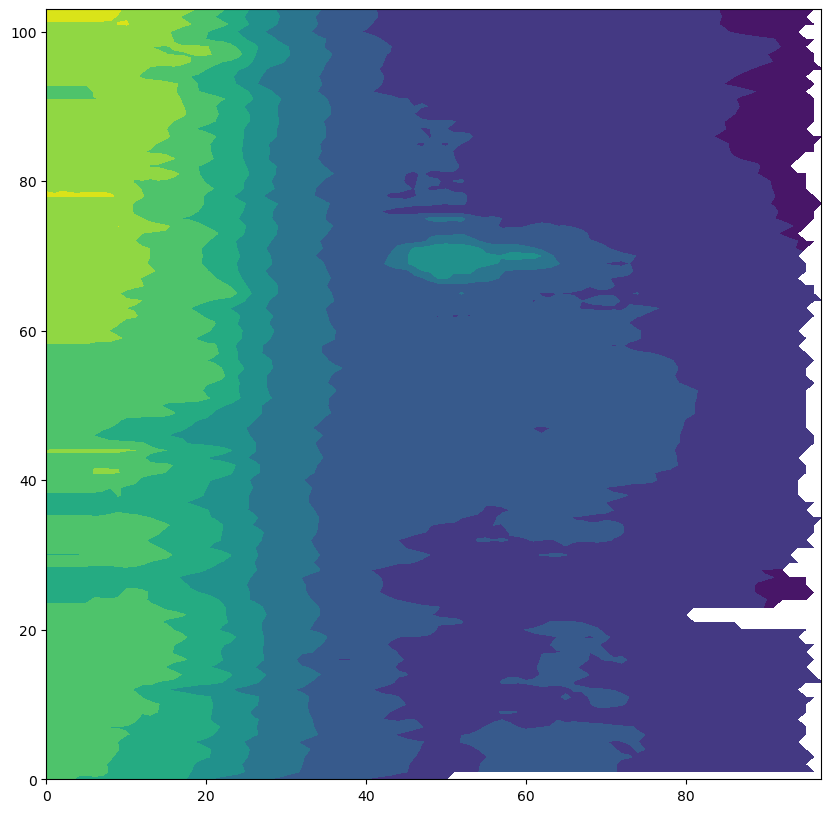
However, we want to add the proper pressure levels since each profile has slightly different levels. For instance note the different values of the pressure observations for profile 3 and 4:
prof.PRES[3,:].values
array([ 6., 7., 8., 9., 10., 16., 25., 36., 46.,
55., 66., 76., 86., 96., 106., 115., 125., 135.,
146., 156., 165., 176., 186., 196., 213., 237., 262.,
288., 313., 338., 363., 388., 413., 438., 463., 488.,
513., 538., 563., 588., 613., 638., 663., 687., 713.,
738., 763., 788., 813., 838., 863., 888., 914., 938.,
963., 988., 1013., 1038., 1063., 1088., 1113., 1139., 1163.,
1188., 1213., 1238., 1263., 1288., 1313., 1338., 1363., 1388.,
1413., 1439., 1463., 1488., 1513., 1538., 1563., 1588., 1613.,
1638., 1663., 1688., 1713., 1738., 1763., 1788., 1813., 1838.,
1863., 1888., 1913., 1938., 1963., 1988., 2013., 2036.],
dtype=float32)
prof.PRES[4,:].values
array([ 6., 7., 8., 9., 10., 16., 25., 35., 45.,
55., 66., 75., 85., 95., 106., 116., 125., 135.,
145., 156., 165., 175., 186., 196., 213., 238., 263.,
287., 313., 338., 363., 388., 413., 438., 463., 488.,
513., 538., 563., 588., 613., 638., 663., 689., 713.,
738., 763., 788., 813., 838., 863., 889., 913., 938.,
963., 988., 1013., 1038., 1063., 1089., 1113., 1138., 1163.,
1188., 1213., 1238., 1263., 1289., 1313., 1338., 1363., 1388.,
1413., 1438., 1462., 1488., 1513., 1538., 1563., 1588., 1613.,
1639., 1663., 1688., 1713., 1737., 1763., 1788., 1813., 1837.,
1863., 1888., 1913., 1938., 1963., 1978., nan, nan],
dtype=float32)
Therefore we need to do a little of interpolation into a common set of pressure values:
prei=np.arange(5,2005,5) # we define a common set of pressure values:
We define the new vectors
juld=prof.JULD.values
psal=prof.PSAL.values
temp=prof.TEMP.values
pres=prof.PRES.values
psali= np.zeros((juld.shape[0],prei.shape[0]))
psali.fill(np.nan)
tempi= np.zeros((juld.shape[0],prei.shape[0]))
tempi.fill(np.nan)
and then we interpolate the salinity and the temperature onto the new levels:
for ip in range(0,pres.shape[0]-1):
psali[ip,:]=np.interp(prei,pres[ip,:],psal[ip,:])
tempi[ip,:]=np.interp(prei,pres[ip,:],temp[ip,:])
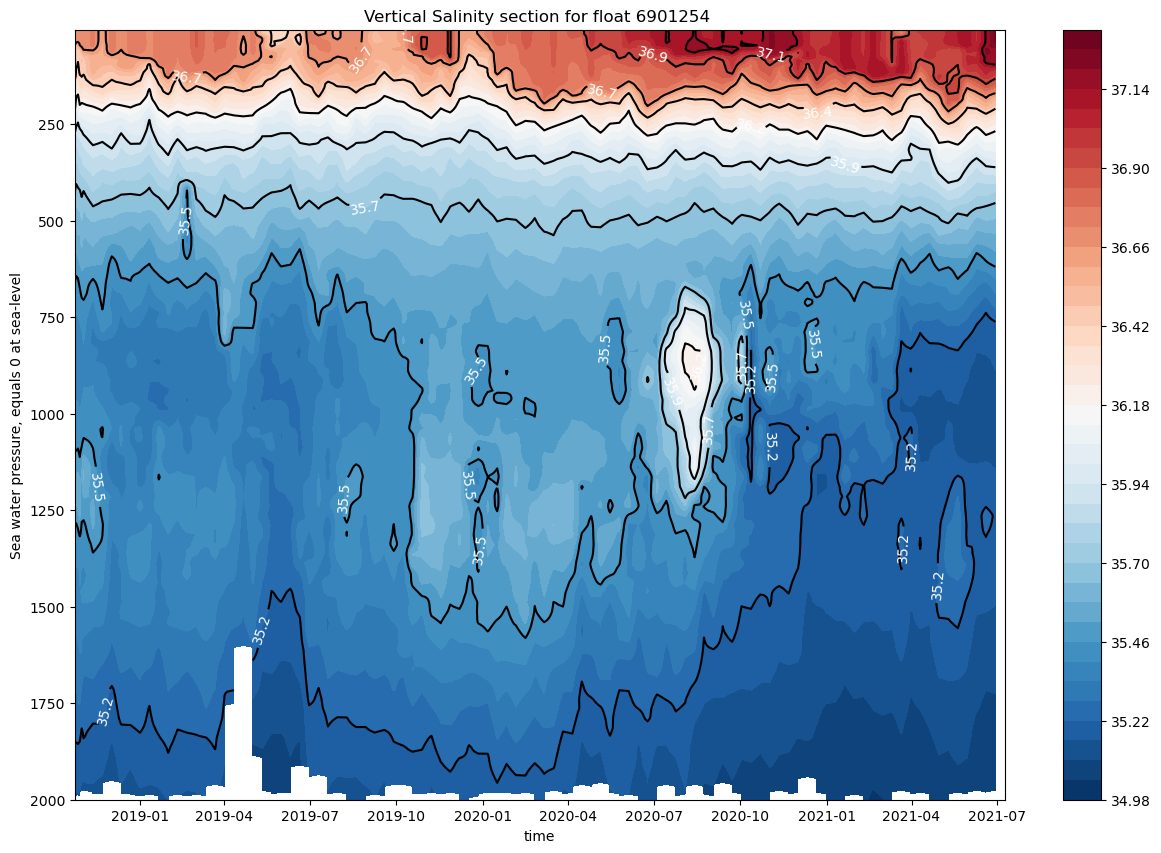
and let’s plot the time evolution of the saliniy measured by this float. in oceanography we usually call it a section
This figure is a little more complex, Unfortunatly we can not teach everything AoS, and therefore we do not explain all the python code, but if you want to get extra information, please make and issue in GitHub
fig, ax = plt.subplots(figsize=(15,10))
#Draw the contours for the salinity
cs=ax.contourf(juld,prei,psali.transpose(),40,cmap="RdBu_r")
#Draw the contours lines to be labelled
cs2=ax.contour(juld,prei,psali.transpose(),colors=('k'), levels=cs.levels[::4])
#Since pressure increase away from the surface we invert the y-axis
ax.invert_yaxis()
ax.clabel(cs2, fmt='%2.1f', colors='w', fontsize=10)
#Add the titles
ax.set_title(f"Vertical Salinity section for float {prof.PLATFORM_NUMBER[0].astype(str).values}")
ax.set_xlabel(f"{prof.JULD.standard_name}")
ax.set_ylabel(f"{prof.PRES.long_name}")
#Add the colorbar
cbar=fig.colorbar(cs,ax=ax)
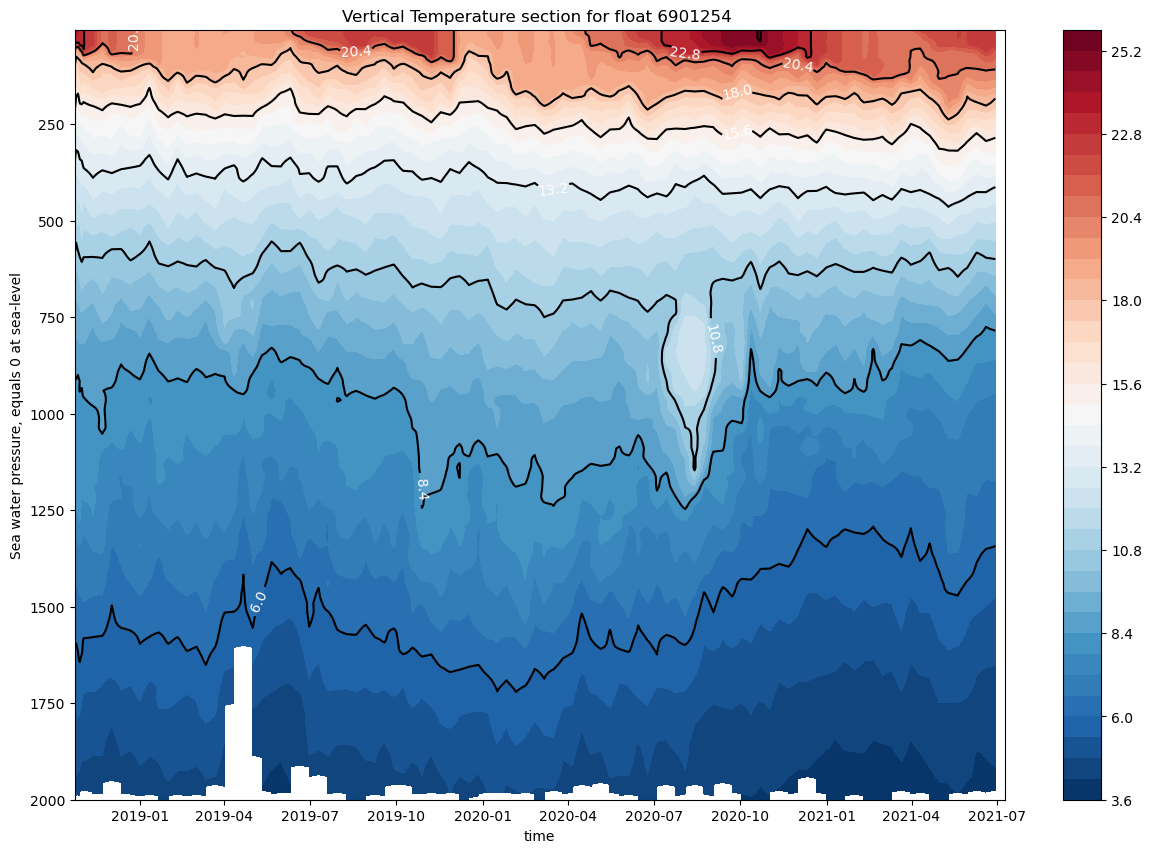
And the same for temperature:
fig, ax = plt.subplots(figsize=(15,10))
cs=ax.contourf(juld,prei,tempi.transpose(),40,cmap="RdBu_r")
cs2=ax.contour(juld,prei,tempi.transpose(),colors=('k'), levels=cs.levels[::4])
ax.invert_yaxis()
ax.clabel(cs2, fmt='%2.1f', colors='w', fontsize=10)
ax.set_title(f"Vertical Temperature section for float {prof.PLATFORM_NUMBER[0].astype(str).values}")
ax.set_xlabel(f"{prof.JULD.standard_name}")
ax.set_ylabel(f"{prof.PRES.long_name}")
cbar=fig.colorbar(cs,ax=ax)
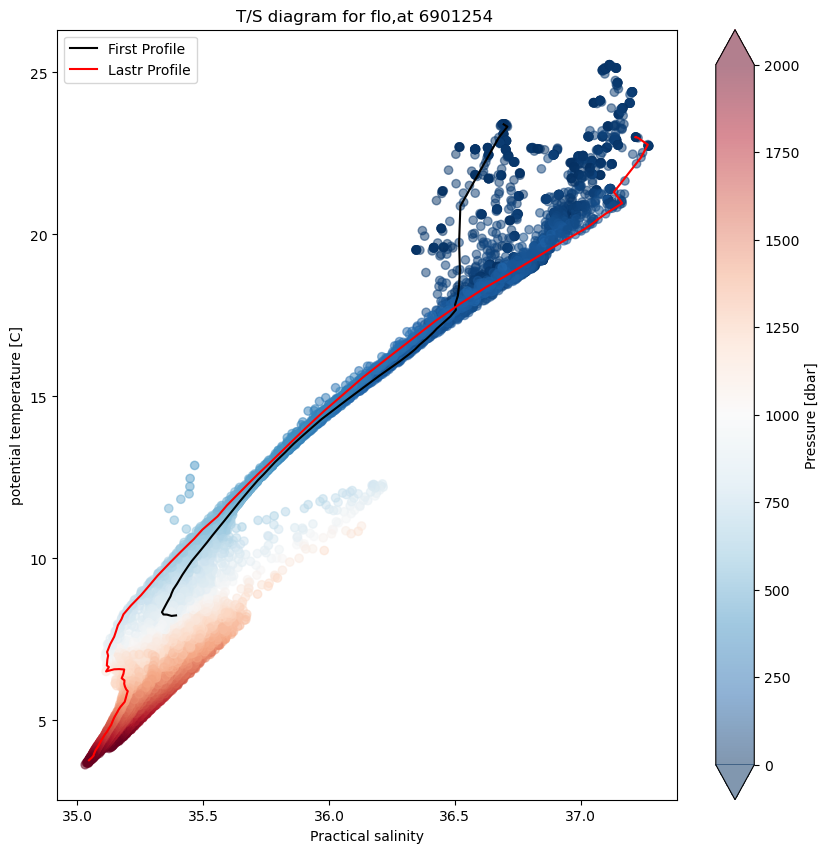
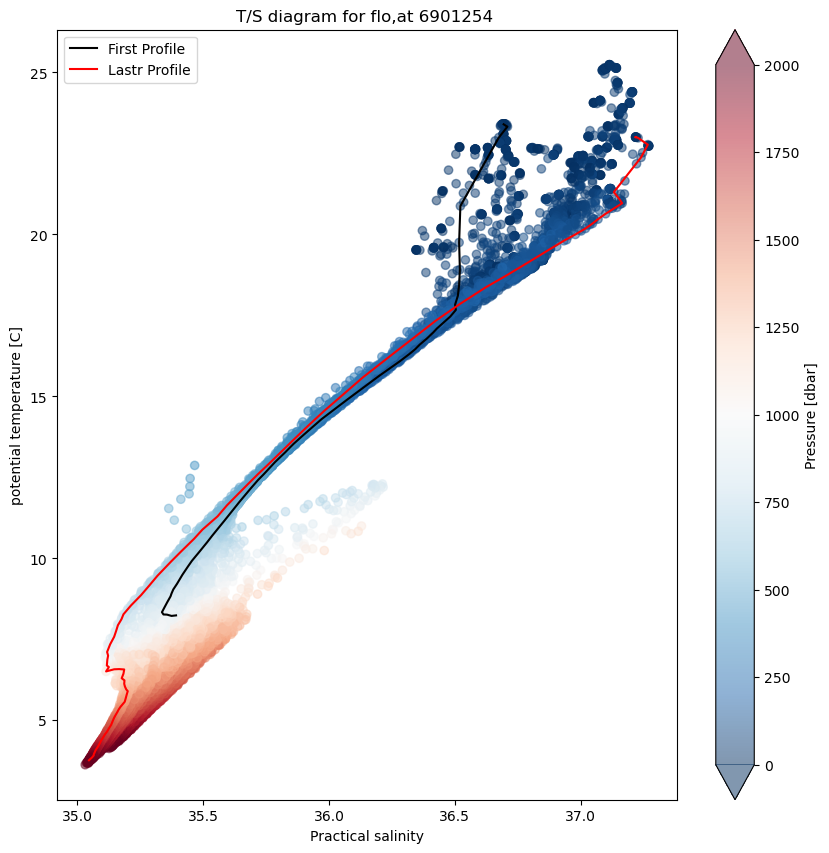
TS-diagram#
In oceanography, temperature-salinity diagrams, sometimes called T-S diagrams, are used to identify water masses. In a T-S diagram, rather than plotting plotting temperatute and salinity as a separate “profile,” with pressure or depth as the vertical coordinate, potential temperature (on the vertical axis) is plotted versus salinity (on the horizontal axis).
This diagrams area very useful since as long as it remains isolated from the surface, where heat or fresh water can be gained or lost, and in the absence of mixing with other water masses, a water parcel’s potential temperature and salinity are conserved. Deep water masses thus retain their T-S characteristics for long periods of time, and can be identified readily on a T-S plot.
In this case we add a colobar bar to show the pressure of each data point.
import seawater as sw
/var/folders/tj/cj2twzcd30jbzn574lsp6phw0000gn/T/ipykernel_1678/1793873256.py:1: UserWarning: The seawater library is deprecated! Please use gsw instead.
import seawater as sw
we will see later how to compute it using gsw
temp=prof.TEMP.values
psal=prof.PSAL.values
pres=prof.PRES.values
ptmp= np.zeros((temp.shape[0],temp.shape[1]))
compute potential temperature
for ip in range(0,temp.shape[0]):
ptmp[ip,:]=sw.ptmp(psal[ip,:], temp[ip,:], pres[ip,:], pr=0)
fig, ax = plt.subplots(figsize=(10,10))
sc=ax.scatter(psal, ptmp, c=pres,alpha=0.5, cmap="RdBu_r",vmin=0, vmax=2000)
ax.plot(psal[0,:], ptmp[0,:],'k',label='First Profile')
ax.plot(psal[-1,:], ptmp[-1,:],'r',label='Lastr Profile')
ax.set_title(f"T/S diagram for flo,at {prof.PLATFORM_NUMBER[0].astype(str).values}")
ax.set_ylabel("potential temperature [C]")
ax.set_xlabel(f"{prof.PSAL.long_name}")
ax.legend()
cbar=fig.colorbar(sc,extend='both');
cbar.set_label('Pressure [dbar]')
import gsw as gsw
long=prof.LONGITUDE.values
lati=prof.LATITUDE.values
ptmp2= np.zeros((temp.shape[0],temp.shape[1]))
sala= np.zeros((temp.shape[0],temp.shape[1]))
for ip in range(0,temp.shape[0]):
sala[ip,:]=gsw.SA_from_SP(psal[ip,:], pres[ip,:], long[ip], lati[ip])
ptmp2[ip,:]=gsw.pt0_from_t(sala[ip,:], temp[ip,:], pres[ip,:])
fig, ax = plt.subplots(figsize=(10,10))
sc=ax.scatter(psal, ptmp2, c=pres,alpha=0.5, cmap="RdBu_r",vmin=0, vmax=2000)
ax.plot(psal[0,:], ptmp2[0,:],'k',label='First Profile')
ax.plot(psal[-1,:], ptmp2[-1,:],'r',label='Lastr Profile')
ax.set_title(f"T/S diagram for flo,at {prof.PLATFORM_NUMBER[0].astype(str).values}")
ax.set_ylabel("potential temperature [C]")
ax.set_xlabel(f"{prof.PSAL.long_name}")
ax.legend()
cbar=fig.colorbar(sc,extend='both');
cbar.set_label('Pressure [dbar]')
Metadata#
And of course, all the metadata information for each profile is included in the netCDF file:
for i1 in range(1,prof.sizes['N_PROF'],10):
print(f"Cycle {prof.data_vars['CYCLE_NUMBER'].values.astype(int)[i1]}"
f" Direction {prof.data_vars['DIRECTION'].values.astype(str)[i1]}"
f" WMO {prof.data_vars['PLATFORM_NUMBER'].values.astype(str)[i1]}"
f" Data Center {prof.data_vars['DATA_CENTRE'].values.astype(str)[i1]}"
f" Project {prof.data_vars['PROJECT_NAME'].values.astype(str)[i1]}" )
Cycle 1 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 11 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 21 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 31 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 41 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 51 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 61 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 71 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 81 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 91 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
Cycle 101 Direction A WMO 6901254 Data Center IF Project ARGO SPAIN
We can see the WMO number, the DAC (IF, for Ifremer) and the owner of the float: Argo Spain
