Delayed mode data#
What if the Real-Time (RT) quality control is not enough?, or if we need the previous data of the float and the data from its neighbors to discern between subtle sensor malfunctioning and natural variability?.
This is where the second data quality control comes in. It is called Delayed - Mode (DM) quality control. Principal investigators and regional data experts are in charge of this meticulous task since it requires a more elaborate specific data analysis of every float and oceanographic expertise in the region where the float was deployed. The most common types of bad data are known as spikes, offsets, drifts, etc… The spikes are abrupt short changes in the data and constitute a clear case of anomaly. Spikes are easy to detect and correct. Drifts and offsets can be identified in the trend of ΔS over time, where ΔS is the difference in salinity between float-measured values and statistical recommendations. If ΔS = a + bt, where t is time, then a is the offset and b is the drift rate. Note that these drifts and offsets can be sensor-related, or they can be due to natural variability, as we explained before. To distinguish between sensor errors and natural variability Evaluation is necessary that experts look carefully at the data. [L2-DM_QC_Graphics]
Delayed mode files#
Delayed mode profile files are the same as the Real-Time profile files, except their file names on the GDACs all contain a “D” before the WMO number (e.g. D5900400_001.nc, BD5904179_001.nc). These profile files contain delayed mode adjusted data, which are recorded in the variable _ADJUSTED. The variable DATA_MODE will record ‘D’. Two other variables are also filled in delayed mode, which are _ADJUSTED_QC and _ADJUSTED_ERROR, which record the delayed mode quality control flags and the delayed mode adjustment uncertainty.
Core Argo delayed mode files are available 1 – 2 years after a profile is taken; sometimes earlier. These have been subjected to detailed scrutiny by oceanographic experts and the adjusted salinity has been estimated by comparison with high-quality ship-based CTD data and Argo climatologies using the process described by Wong et al, 2003; Böhme and Send, 2005; Owens and Wong, 2009; Cabanes et al, 2016.
For BGC parameters, delayed mode files can be available within 5 – 6 cycles after deployment. This is because the BGC sensors often return data that are out of calibration, but early adjustment methodologies exist that can significantly improve their accuracy. Additional delayed mode quality control occurs when a longer record of float data is available.
To learn more about delayed mode quality control, read the papers on the methods linked above or see the ADMT documentation page for core Argo and BGC-Argo quality control documentation.
For each parameter, there are two variables associated with it: a raw version and an adjusted version. The raw version can be found in the “PARAM” variable (e.g. TEMP, PRES, DOXY) and the adjusted version can be found in the “PARAM_ADJUSTED” variable (e.g. TEMP_ADJUSTED, PRES_ADJUSTED, DOXY_ADJUSTED).
Quality Control flags#
QCflag |
Meaning |
Real time description |
Adjusted description |
|---|---|---|---|
0 |
No QC performed |
No QC performed |
XX |
1 |
Good data |
All real time QC tests passed |
XX |
2 |
Probably good data |
Probably good |
XX |
3 |
Bad data that are potentially correctable |
Test 15 or Test 16 or Test 17 failed and all other real-time QC tests passed. These data are not to be used without scientific correction. A flag ‘3’ may be assigned by an operator during additional visual QC for bad |
data that may be corrected in delayed mode. |
4 |
Bad data |
Data have failed one or more of the real-time QC tests, excluding Test 16. A flag ‘4’ may be assigned by an operator during additional visual QC for bad data that are not correctable. |
XX |
5 |
Value changed |
Value changed |
XX |
6 |
Not currently used |
Not currently used |
XX |
7 |
Not currently used |
Not currently used |
XX |
8 |
Estimated |
Estimated value (interpolated, extrapolated or other estimation) |
XX |
9 |
Missing value |
Missing value |
XX |
Example#
import numpy as np
import netCDF4
import xarray as xr
import cartopy.crs as ccrs
import cartopy
import matplotlib as mpl
import matplotlib.cm as cm
from matplotlib import pyplot as plt
%matplotlib inline
qcmap = mpl.colors.ListedColormap(['#000000' , '#31FC03' , '#ADFC03' , '#FCBA03' ,'#FC1C03',
'#324CA8' , '#000000' , '#000000' , '#B22CC9', '#000000'])
def colorbar_qc(cmap, **kwargs):
"""Adjust colorbar ticks with discrete colors for QC flags"""
ncolors = 10
mappable = cm.ScalarMappable(cmap=cmap)
mappable.set_array([])
mappable.set_clim(-0.5, ncolors+0.5)
colorbar = plt.colorbar(mappable, **kwargs)
colorbar.set_ticks(np.linspace(0, ncolors, ncolors))
colorbar.set_ticklabels(range(ncolors))
return colorbar
iwmo=6900768
file=f"/Users/pvb/Dropbox/Oceanografia/Data/Argo/Floats/{iwmo}/{iwmo}_prof.nc"
prof = xr.open_dataset(file)
All data, with any of the QC flags
fig, ax = plt.subplots(figsize=(15,8))
sc = ax.scatter(prof.PSAL,prof.TEMP,c=prof.CYCLE_NUMBER+prof.PRES*0)
cbar=fig.colorbar(sc,ax=ax)
ax.set_title(f"{iwmo}")
ax.grid()
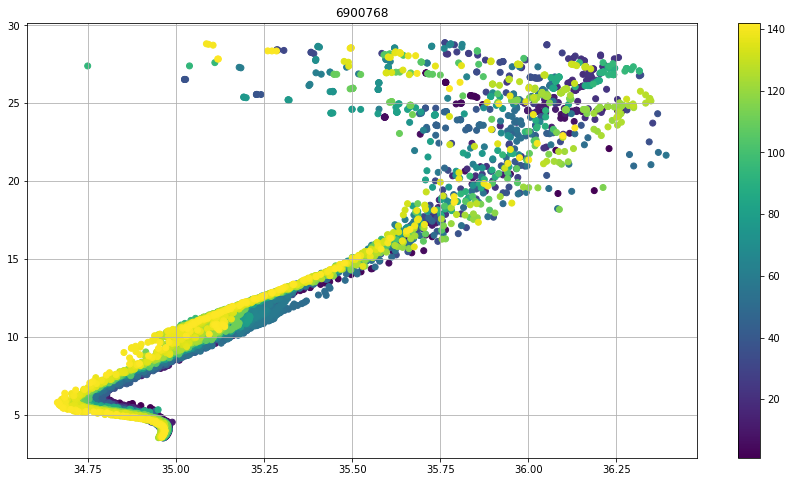
psal=prof.PSAL.where(prof.PSAL_QC.values.astype(float) == 2)
temp=prof.TEMP.where(prof.PSAL_QC.values.astype(float) == 2)
fig, ax = plt.subplots(figsize=(15,8))
sc = ax.scatter(psal,temp,c=prof.CYCLE_NUMBER+prof.PRES*0)
cbar=fig.colorbar(sc,ax=ax)
ax.set_title(f"{iwmo}")
ax.grid()
psal
<xarray.DataArray 'PSAL' (N_PROF: 142, N_LEVELS: 71)>
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]], dtype=float32)
Dimensions without coordinates: N_PROF, N_LEVELS
Attributes:
long_name: Practical salinity
standard_name: sea_water_salinity
units: psu
valid_min: 2.0
valid_max: 41.0
C_format: %9.3f
FORTRAN_format: F9.3
resolution: 0.001- N_PROF: 142
- N_LEVELS: 71
- nan nan nan nan nan nan nan nan ... nan nan nan nan nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - long_name :
- Practical salinity
- standard_name :
- sea_water_salinity
- units :
- psu
- valid_min :
- 2.0
- valid_max :
- 41.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001

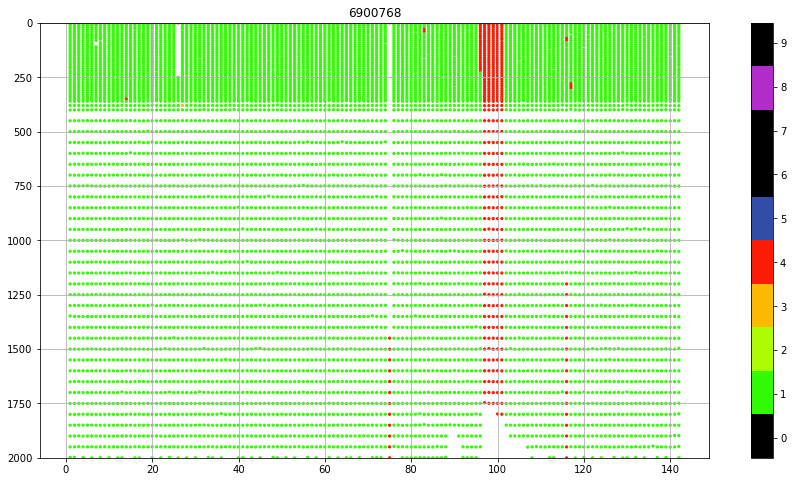
fig, ax = plt.subplots(figsize=(15,8))
sc = ax.scatter(prof.CYCLE_NUMBER+prof.PRES*0,
prof.PRES,
c=prof.PSAL_QC, vmin=0, vmax=9, cmap=qcmap, s=5)
colorbar_qc(qcmap, ax=ax)
ax.set_title(f"{iwmo}")
ax.grid()
ax.set_ylim(0,2000)
ax.invert_yaxis()
prof.PSAL[90,:]
<xarray.DataArray 'PSAL' (N_LEVELS: 71)>
array([36.213, 36.213, 36.209, 36.208, 36.204, 36.133, 35.764, 35.613, 35.538,
35.447, 35.398, 35.358, 35.329, 35.317, 35.285, 35.247, 35.228, 35.211,
35.191, 35.164, 35.154, 35.129, 35.112, 35.092, 35.082, 35.071, 35.061,
35.046, 35.037, 35.032, 35.028, 35.023, 35.056, 35.062, 35.048, 35.026,
35.031, 35.036, 35.028, 35.017, 34.981, 34.927, 34.921, 34.858, 34.814,
34.777, 34.739, 34.72 , 34.722, 34.733, 34.756, 34.795, 34.816, 34.855,
34.886, 34.904, 34.918, 34.942, 34.955, 34.965, 34.971, 34.975, 34.977,
34.976, 34.976, 34.975, 34.974, 34.973, 34.971, nan, nan],
dtype=float32)
Dimensions without coordinates: N_LEVELS
Attributes:
long_name: Practical salinity
standard_name: sea_water_salinity
units: psu
valid_min: 2.0
valid_max: 41.0
C_format: %9.3f
FORTRAN_format: F9.3
resolution: 0.001- N_LEVELS: 71
- 36.21 36.21 36.21 36.21 36.2 36.13 ... 34.97 34.97 34.97 34.97 nan nan
array([36.213, 36.213, 36.209, 36.208, 36.204, 36.133, 35.764, 35.613, 35.538, 35.447, 35.398, 35.358, 35.329, 35.317, 35.285, 35.247, 35.228, 35.211, 35.191, 35.164, 35.154, 35.129, 35.112, 35.092, 35.082, 35.071, 35.061, 35.046, 35.037, 35.032, 35.028, 35.023, 35.056, 35.062, 35.048, 35.026, 35.031, 35.036, 35.028, 35.017, 34.981, 34.927, 34.921, 34.858, 34.814, 34.777, 34.739, 34.72 , 34.722, 34.733, 34.756, 34.795, 34.816, 34.855, 34.886, 34.904, 34.918, 34.942, 34.955, 34.965, 34.971, 34.975, 34.977, 34.976, 34.976, 34.975, 34.974, 34.973, 34.971, nan, nan], dtype=float32) - long_name :
- Practical salinity
- standard_name :
- sea_water_salinity
- units :
- psu
- valid_min :
- 2.0
- valid_max :
- 41.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
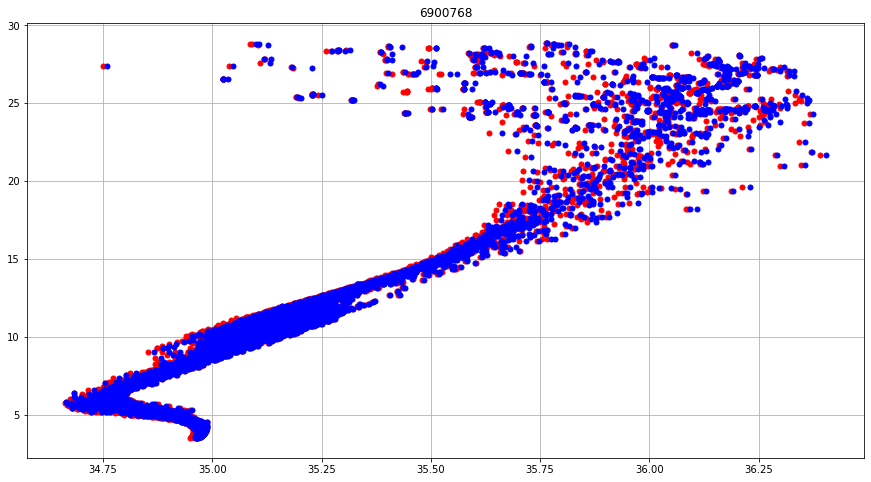
fig, ax = plt.subplots(figsize=(15,8))
scRT = ax.plot(prof.PSAL, prof.TEMP, 'ro', markersize=5)
scAD = ax.plot(prof.PSAL_ADJUSTED, prof.TEMP_ADJUSTED, 'bo',markersize=5)
ax.set_title(f"{iwmo}")
ax.grid()
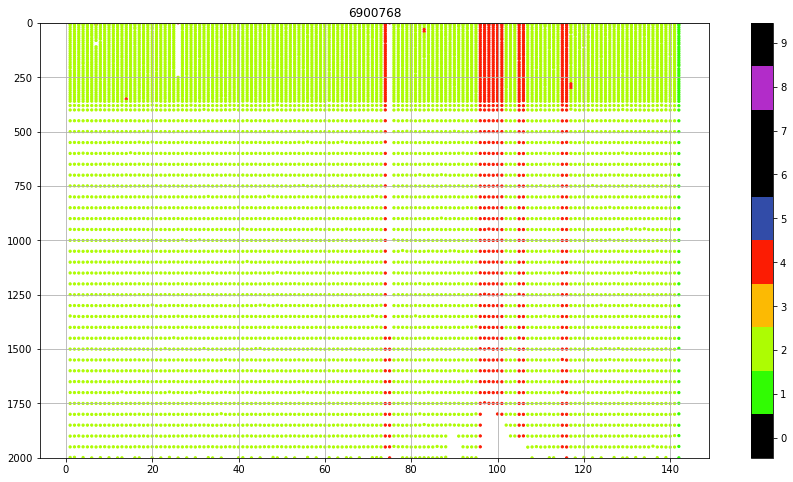
fig, ax = plt.subplots(figsize=(15,8))
sc = ax.scatter(prof.CYCLE_NUMBER+prof.PRES*0,
prof.PRES_ADJUSTED,
c=prof.PSAL_ADJUSTED_QC, vmin=0, vmax=9, cmap=qcmap, s=5)
colorbar_qc(qcmap, ax=ax)
ax.set_title(f"{iwmo}")
ax.grid()
ax.set_ylim(0,2000)
ax.invert_yaxis()
prof
<xarray.Dataset>
Dimensions: (N_PROF: 142, N_PARAM: 3, N_LEVELS: 71, N_CALIB: 1, N_HISTORY: 0)
Dimensions without coordinates: N_PROF, N_PARAM, N_LEVELS, N_CALIB, N_HISTORY
Data variables: (12/64)
DATA_TYPE object b'Argo profile '
FORMAT_VERSION object b'3.1 '
HANDBOOK_VERSION object b'1.2 '
REFERENCE_DATE_TIME object b'19500101000000'
DATE_CREATION object b'20110107224755'
DATE_UPDATE object b'20181120184739'
... ...
HISTORY_ACTION (N_HISTORY, N_PROF) object
HISTORY_PARAMETER (N_HISTORY, N_PROF) object
HISTORY_START_PRES (N_HISTORY, N_PROF) float32
HISTORY_STOP_PRES (N_HISTORY, N_PROF) float32
HISTORY_PREVIOUS_VALUE (N_HISTORY, N_PROF) float32
HISTORY_QCTEST (N_HISTORY, N_PROF) object
Attributes:
title: Argo float vertical profile
institution: FR GDAC
source: Argo float
history: 2018-11-20T18:47:39Z creation
references: http://www.argodatamgt.org/Documentation
user_manual_version: 3.1
Conventions: Argo-3.1 CF-1.6
featureType: trajectoryProfile- N_PROF: 142
- N_PARAM: 3
- N_LEVELS: 71
- N_CALIB: 1
- N_HISTORY: 0
- DATA_TYPE()object...
- long_name :
- Data type
- conventions :
- Argo reference table 1
array(b'Argo profile ', dtype=object)
- FORMAT_VERSION()object...
- long_name :
- File format version
array(b'3.1 ', dtype=object)
- HANDBOOK_VERSION()object...
- long_name :
- Data handbook version
array(b'1.2 ', dtype=object)
- REFERENCE_DATE_TIME()object...
- long_name :
- Date of reference for Julian days
- conventions :
- YYYYMMDDHHMISS
array(b'19500101000000', dtype=object)
- DATE_CREATION()object...
- long_name :
- Date of file creation
- conventions :
- YYYYMMDDHHMISS
array(b'20110107224755', dtype=object)
- DATE_UPDATE()object...
- long_name :
- Date of update of this file
- conventions :
- YYYYMMDDHHMISS
array(b'20181120184739', dtype=object)
- PLATFORM_NUMBER(N_PROF)object...
- long_name :
- Float unique identifier
- conventions :
- WMO float identifier : A9IIIII
array([b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 ', b'6900768 '], dtype=object) - PROJECT_NAME(N_PROF)object...
- long_name :
- Name of the project
array([b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN ', b'ARGO_SPAIN '], dtype=object) - PI_NAME(N_PROF)object...
- long_name :
- Name of the principal investigator
array([b'Pedro Joaquin VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI ', b'Pedro VELEZ BELCHI '], dtype=object) - STATION_PARAMETERS(N_PROF, N_PARAM)object...
- long_name :
- List of available parameters for the station
- conventions :
- Argo reference table 3
array([[b'PRES ', b'TEMP ', b'PSAL '], [b'PRES ', b'TEMP ', b'PSAL '], [b'PRES ', b'TEMP ', b'PSAL '], ..., [b'PRES ', b'TEMP ', b'PSAL '], [b'PRES ', b'TEMP ', b'PSAL '], [b'PRES ', b'TEMP ', b'PSAL ']], dtype=object) - CYCLE_NUMBER(N_PROF)float64...
- long_name :
- Float cycle number
- conventions :
- 0...N, 0 : launch cycle (if exists), 1 : first complete cycle
array([ 1., 2., 3., 4., 5., 6., 7., 8., 9., 10., 11., 12., 13., 14., 15., 16., 17., 18., 19., 20., 21., 22., 23., 24., 25., 26., 27., 28., 29., 30., 31., 32., 33., 34., 35., 36., 37., 38., 39., 40., 41., 42., 43., 44., 45., 46., 47., 48., 49., 50., 51., 52., 53., 54., 55., 56., 57., 58., 59., 60., 61., 62., 63., 64., 65., 66., 67., 68., 69., 70., 71., 72., 73., 74., 75., 76., 77., 78., 79., 80., 81., 82., 83., 84., 85., 86., 87., 88., 89., 90., 91., 92., 93., 94., 95., 96., 97., 98., 99., 100., 101., 102., 103., 104., 105., 106., 107., 108., 109., 110., 111., 112., 113., 114., 115., 116., 117., 118., 119., 120., 121., 122., 123., 124., 125., 126., 127., 128., 129., 130., 131., 132., 133., 134., 135., 136., 137., 138., 139., 140., 141., 142.]) - DIRECTION(N_PROF)object...
- long_name :
- Direction of the station profiles
- conventions :
- A: ascending profiles, D: descending profiles
array([b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A'], dtype=object) - DATA_CENTRE(N_PROF)object...
- long_name :
- Data centre in charge of float data processing
- conventions :
- Argo reference table 4
array([b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF'], dtype=object) - DC_REFERENCE(N_PROF)object...
- long_name :
- Station unique identifier in data centre
- conventions :
- Data centre convention
array([b' ', b'15347391 ', b'15425524 ', b'15527281 ', b'15601720 ', b'15702801 ', b'15961520 ', b'16440641 ', b'16761583 ', b'16834351 ', b'18148044 ', b'18546933 ', b'18589206 ', b'18647267 ', b'18725425 ', b'22807853 ', b'23341001 ', b'23437103 ', b'23498483 ', b'23630036 ', b'23699985 ', b'23788424 ', b'23859645 ', b'23929106 ', b'24003565 ', b'24074288 ', b'24165544 ', b'24237225 ', b'24330083 ', b'24402423 ', b'24635247 ', b'24714366 ', b'24811061 ', b'24893441 ', b'24959544 ', b'25041205 ', b'25109043 ', b'25156703 ', b'25255404 ', b'25306509 ', b'25384589 ', b'25446096 ', b'25533446 ', b'25594510 ', b'25655128 ', b'25725647 ', b'25767526 ', b'25851595 ', b'25911074 ', b'25994187 ', b'26062268 ', b'26163410 ', b'26215448 ', b'26282130 ', b'26374047 ', b'26448028 ', b'26534767 ', b'26584526 ', b'26677246 ', b'26813108 ', b'26881718 ', b'27019867 ', b'27327806 ', b'27433868 ', b'27522606 ', b'27588586 ', b'27684706 ', b'27749327 ', b'27840679 ', b'27924469 ', b'28016009 ', b'28099452 ', b'28168769 ', b'28245552 ', b'28304363 ', b'28402410 ', b'28472530 ', b'28549572 ', b'28621115 ', b'28693969 ', b'28783119 ', b'28841215 ', b'29154069 ', b'29137389 ', b'29236612 ', b'29309609 ', b'29381409 ', b'29471710 ', b'29537158 ', b'29627489 ', b'29710101 ', b'29806389 ', b'29887409 ', b'29971691 ', b'30080809 ', b'30162810 ', b'30267529 ', b'30368829 ', b'30446389 ', b'30542650 ', b'30645106 ', b'30745529 ', b'30859117 ', b'30955111 ', b'31039449 ', b'31121872 ', b'31337689 ', b'31741530 ', b'31835510 ', b'31918749 ', b'32007500 ', b'32087632 ', b'32182930 ', b'32254849 ', b'32334569 ', b'32433998 ', b'32508970 ', b'32595909 ', b'32661123 ', b'32736727 ', b'32814549 ', b'32873055 ', b'32978109 ', b'33040916 ', b'33127038 ', b'33189569 ', b'33273438 ', b'33465382 ', b'33714424 ', b'33731214 ', b'33738531 ', b'33850625 ', b'33924784 ', b'34009357 ', b'34082856 ', b'34158157 ', b'34252911 ', b'34317677 ', b'34403871 ', b'34473498 ', b'34592326 ', b'35039559 '], dtype=object) - DATA_STATE_INDICATOR(N_PROF)object...
- long_name :
- Degree of processing the data have passed through
- conventions :
- Argo reference table 6
array([b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C '], dtype=object) - DATA_MODE(N_PROF)object...
- long_name :
- Delayed mode or real time data
- conventions :
- R : real time; D : delayed mode; A : real time with adjustment
array([b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D', b'D'], dtype=object) - PLATFORM_TYPE(N_PROF)object...
- long_name :
- Type of float
- conventions :
- Argo reference table 23
array([b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX ', b'APEX '], dtype=object) - FLOAT_SERIAL_NO(N_PROF)object...
- long_name :
- Serial number of the float
array([b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 ', b'4403 '], dtype=object) - FIRMWARE_VERSION(N_PROF)object...
- long_name :
- Instrument firmware version
array([b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 ', b'062608 '], dtype=object) - WMO_INST_TYPE(N_PROF)object...
- long_name :
- Coded instrument type
- conventions :
- Argo reference table 8
array([b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 ', b'846 '], dtype=object) - JULD(N_PROF)datetime64[ns]...
- long_name :
- Julian day (UTC) of the station relative to REFERENCE_DATE_TIME
- standard_name :
- time
- conventions :
- Relative julian days with decimal part (as parts of day)
- resolution :
- 0.0
- axis :
- T
array(['2010-12-28T10:48:04.000000000', '2011-01-07T10:37:48.000000000', '2011-01-17T13:15:26.000000000', '2011-01-27T10:40:17.000000000', '2011-02-06T12:52:59.000000000', '2011-02-16T10:55:14.000000000', '2011-02-26T12:58:52.000000000', '2011-03-08T10:25:59.000000000', '2011-03-18T13:07:35.000000000', '2011-03-28T10:12:35.000000000', '2011-04-07T13:15:09.000000000', '2011-04-17T10:33:43.999999744', '2011-04-27T12:43:02.000000000', '2011-05-07T13:07:30.000000000', '2011-05-17T13:10:27.000000000', '2011-05-27T10:40:57.000000000', '2011-06-06T10:35:23.000000000', '2011-06-16T13:10:46.000000000', '2011-06-26T13:13:42.000000000', '2011-07-06T10:57:15.000000000', '2011-07-16T13:19:01.000000000', '2011-07-26T10:47:49.000000000', '2011-08-05T13:12:26.000000000', '2011-08-15T10:54:50.000000000', '2011-08-25T13:15:28.000000256', '2011-09-04T11:02:57.000000000', '2011-09-14T13:15:05.000000000', '2011-09-24T11:09:56.000000000', '2011-10-04T13:07:54.000000000', '2011-10-14T11:27:41.000000000', '2011-10-24T13:25:05.000000000', '2011-11-03T13:18:57.000000000', '2011-11-13T10:45:30.000000000', '2011-11-23T11:16:46.000000000', '2011-12-03T13:14:44.999999744', '2011-12-13T11:18:07.000000000', '2011-12-23T13:18:55.000000256', '2012-01-02T13:22:25.000000000', '2012-01-12T13:15:42.000000000', '2012-01-22T10:36:34.000000000', '2012-02-01T10:34:58.000000000', '2012-02-11T13:10:20.000000256', '2012-02-21T10:35:44.000000000', '2012-03-02T13:05:59.000000000', '2012-03-12T11:11:36.000000000', '2012-03-22T13:12:58.000000000', '2012-04-01T13:11:55.000000000', '2012-04-11T10:32:13.000000000', '2012-04-21T11:15:57.000000000', '2012-05-01T13:08:49.000000000', '2012-05-11T11:20:07.000000000', '2012-05-21T13:15:14.000000000', '2012-05-31T12:36:48.000000000', '2012-06-10T13:06:21.000000000', '2012-06-20T13:06:27.000000000', '2012-06-30T10:43:09.999999744', '2012-07-10T13:24:11.000000256', '2012-07-20T11:06:35.000000256', '2012-07-30T13:20:24.000000000', '2012-08-09T13:34:06.000000000', '2012-08-19T10:46:27.000000000', '2012-08-29T13:16:09.000000000', '2012-09-08T11:12:08.000000000', '2012-09-18T13:28:48.000000256', '2012-09-28T11:10:03.000000000', '2012-10-08T11:00:31.000000000', '2012-10-18T13:30:12.000000000', '2012-10-28T11:16:33.000000000', '2012-11-07T13:27:33.000000000', '2012-11-17T13:26:30.999999744', '2012-11-27T10:41:43.000000000', '2012-12-07T10:40:07.000000256', '2012-12-17T11:05:10.000000000', '2012-12-27T13:25:12.999999744', '2013-01-06T11:04:13.000000000', '2013-01-16T13:11:49.000000000', '2013-01-26T13:27:47.000000000', '2013-02-05T10:36:45.000000000', '2013-02-15T10:04:32.000000000', '2013-02-25T09:40:50.000000000', '2013-03-07T09:40:22.000000000', '2013-03-17T10:16:11.000000256', '2013-03-27T09:51:21.000000000', '2013-04-06T10:02:13.000000000', '2013-04-16T09:49:52.000000000', '2013-04-26T09:57:20.000000000', '2013-05-06T10:08:45.999999744', '2013-05-16T10:56:28.000000000', '2013-05-26T13:08:01.000000000', '2013-06-05T13:06:59.000000000', '2013-06-15T13:12:10.000000000', '2013-06-25T10:21:07.000000000', '2013-07-05T10:14:25.000000000', '2013-07-15T09:47:52.000000000', '2013-07-25T10:17:26.000000000', '2013-08-04T13:25:05.000000000', '2013-08-14T12:43:49.000000000', '2013-08-24T12:42:12.000000000', '2013-09-03T12:57:36.000000000', '2013-09-13T12:43:31.000000000', '2013-09-23T13:02:53.000000000', '2013-10-03T13:09:12.000000000', '2013-10-13T13:18:21.000000000', '2013-10-23T13:16:11.000000256', '2013-11-02T13:11:44.000000000', '2013-11-12T13:27:08.000000000', '2013-11-22T13:21:33.000000000', '2013-12-02T12:53:19.000000000', '2013-12-12T13:18:54.000000000', '2013-12-22T13:12:45.000000000', '2014-01-01T13:06:03.000000000', '2014-01-11T10:15:35.000000000', '2014-01-21T11:04:58.000000000', '2014-01-31T13:18:48.000000000', '2014-02-10T13:27:24.000000000', '2014-02-20T11:07:31.000000000', '2014-03-02T13:14:32.000000000', '2014-03-12T10:30:18.000000000', '2014-03-22T13:13:01.000000000', '2014-04-01T10:39:31.000000000', '2014-04-11T13:25:39.000000000', '2014-04-21T11:43:10.000000000', '2014-05-01T13:24:08.000000000', '2014-05-11T11:55:15.000000000', '2014-05-21T13:21:28.000000000', '2014-05-31T13:25:32.000000000', '2014-06-10T10:57:43.000000000', '2014-06-20T13:26:50.000000000', '2014-06-30T10:52:48.000000000', '2014-07-10T13:24:11.000000256', '2014-07-20T13:29:23.000000256', '2014-07-30T11:15:10.000000256', '2014-08-09T13:34:39.000000256', '2014-08-19T11:21:35.000000000', '2014-08-29T13:27:28.000000000', '2014-09-08T13:23:02.000000000', '2014-09-18T10:58:37.000000000', '2014-09-28T13:36:49.000000000', '2014-10-08T11:21:27.999999744', '2014-10-18T13:33:36.000000000', '2014-10-28T13:43:19.000000000', '2014-11-07T11:38:45.000000000'], dtype='datetime64[ns]') - JULD_QC(N_PROF)object...
- long_name :
- Quality on date and time
- conventions :
- Argo reference table 2
array([b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1'], dtype=object) - JULD_LOCATION(N_PROF)datetime64[ns]...
- long_name :
- Julian day (UTC) of the location relative to REFERENCE_DATE_TIME
- conventions :
- Relative julian days with decimal part (as parts of day)
- resolution :
- 0.0
array(['2010-12-28T11:52:27.000000000', '2011-01-07T11:17:49.000000000', '2011-01-17T13:43:51.000000000', '2011-01-27T11:49:29.000000000', '2011-02-06T13:36:23.999999744', '2011-02-16T11:46:35.000000000', '2011-02-26T13:29:49.000000000', '2011-03-08T11:12:13.999999744', '2011-03-18T15:03:15.000000000', '2011-03-28T10:26:21.000000000', '2011-04-07T14:55:47.999999744', '2011-04-17T11:08:56.000000000', '2011-04-27T14:49:11.000000000', '2011-05-07T14:46:44.000000000', '2011-05-17T14:42:53.000000000', '2011-05-27T11:36:16.000000000', '2011-06-06T11:02:23.000000000', '2011-06-16T14:31:14.000000000', '2011-06-26T14:27:11.000000000', '2011-07-06T11:47:28.000000000', '2011-07-16T14:14:20.000000000', '2011-07-26T11:24:26.000000000', '2011-08-05T14:14:44.000000000', '2011-08-15T11:05:03.000000000', '2011-08-25T14:02:26.000000000', '2011-09-04T11:13:46.000000000', '2011-09-14T13:56:13.999999744', '2011-09-24T12:34:44.000000000', '2011-10-04T13:49:03.000000000', '2011-10-14T12:19:54.000000000', '2011-10-24T13:38:28.000000000', '2011-11-03T13:33:28.000000000', '2011-11-13T11:57:32.000000000', '2011-11-23T11:51:07.000000000', '2011-12-03T15:05:02.000000000', '2011-12-13T11:37:10.000000000', '2011-12-23T14:54:19.000000000', '2012-01-02T14:49:19.000000000', '2012-01-12T14:45:01.000000256', '2012-01-22T10:54:24.000000000', '2012-02-01T10:46:39.000000000', '2012-02-11T14:29:44.000000256', '2012-02-21T10:51:00.000000000', '2012-03-02T14:19:26.000000000', '2012-03-12T12:17:42.000000000', '2012-03-22T14:08:50.999999744', '2012-04-01T14:01:00.000000000', '2012-04-11T11:55:51.999999744', '2012-04-21T11:28:29.000000000', '2012-05-01T13:42:12.000000000', '2012-05-11T11:35:12.000000000', '2012-05-21T13:32:18.000000000', '2012-05-31T13:07:27.999999744', '2012-06-10T14:54:45.000000000', '2012-06-20T14:45:23.999999744', '2012-06-30T10:59:40.000000000', '2012-07-10T14:47:51.000000000', '2012-07-20T11:53:41.000000000', '2012-07-30T14:37:32.000000256', '2012-08-09T14:31:24.000000000', '2012-08-19T10:57:51.000000000', '2012-08-29T14:17:47.000000000', '2012-09-08T11:52:27.000000000', '2012-09-18T14:05:08.000000000', '2012-09-28T11:38:44.000000000', '2012-10-08T11:31:45.000000256', '2012-10-18T13:44:04.000000000', '2012-10-28T11:53:53.000000000', '2012-11-07T16:02:37.000000000', '2012-11-17T15:51:56.000000000', '2012-11-27T10:56:48.000000000', '2012-12-07T10:50:47.000000000', '2012-12-17T11:50:51.000000000', '2012-12-27T14:42:04.999999744', '2013-01-06T11:15:23.000000000', '2013-01-16T14:27:18.000000000', '2013-01-26T14:25:05.000000000', '2013-02-05T11:47:55.999999744', '2013-02-15T11:27:04.000000256', '2013-02-25T09:54:13.000000000', '2013-03-07T09:52:20.000000000', '2013-03-17T10:29:56.000000000', '2013-03-27T10:05:26.999999744', '2013-04-06T11:06:53.000000000', '2013-04-16T10:59:55.000000000', '2013-04-26T10:45:44.000000000', '2013-05-06T10:25:33.000000256', '2013-05-16T11:40:44.000000000', '2013-05-26T14:41:17.999999744', '2013-06-05T14:33:45.000000000', '2013-06-15T14:25:37.000000000', '2013-06-25T11:07:57.000000256', '2013-07-05T11:00:57.000000000', '2013-07-15T10:02:40.000000000', '2013-07-25T10:47:06.000000000', '2013-08-04T15:28:39.000000256', '2013-08-14T12:59:11.000000000', '2013-08-24T12:52:44.000000000', '2013-09-03T15:05:27.000000000', '2013-09-13T15:00:09.000000000', '2013-09-23T14:52:19.000000000', '2013-10-03T14:45:02.000000000', '2013-10-13T14:35:30.000000000', '2013-10-23T14:27:29.000000000', '2013-11-02T14:19:30.999999744', '2013-11-12T14:10:50.000000000', '2013-11-22T14:01:51.000000000', '2013-12-02T13:27:28.000000000', '2013-12-12T15:28:03.000000000', '2013-12-22T15:19:23.000000000', '2014-01-01T15:10:20.000000000', '2014-01-11T10:31:03.000000000', '2014-01-21T11:17:33.999999744', '2014-01-31T14:46:41.999999744', '2014-02-10T14:36:23.000000000', '2014-02-20T11:44:18.000000256', '2014-03-02T14:19:50.000000000', '2014-03-12T10:42:33.000000000', '2014-03-22T14:00:23.999999744', '2014-04-01T11:13:54.000000000', '2014-04-11T15:24:16.999999744', '2014-04-21T12:40:28.000000000', '2014-05-01T15:06:12.000000000', '2014-05-11T12:06:26.000000000', '2014-05-21T14:47:35.000000000', '2014-05-31T14:40:58.000000000', '2014-06-10T11:10:32.000000000', '2014-06-20T14:13:59.000000000', '2014-06-30T11:06:33.000000000', '2014-07-10T15:41:40.000000000', '2014-07-20T15:31:22.000000000', '2014-07-30T11:31:57.000000000', '2014-08-09T15:12:12.000000000', '2014-08-19T12:03:51.999999744', '2014-08-29T14:52:34.000000000', '2014-09-08T14:42:26.000000000', '2014-09-18T11:42:19.000000000', '2014-09-28T14:18:12.000000000', '2014-10-08T13:13:11.000000000', '2014-10-18T15:44:36.000000000', '2014-10-28T15:33:46.000000000', '2014-11-07T12:02:32.000000000'], dtype='datetime64[ns]') - LATITUDE(N_PROF)float64...
- long_name :
- Latitude of the station, best estimate
- standard_name :
- latitude
- units :
- degree_north
- valid_min :
- -90.0
- valid_max :
- 90.0
- axis :
- Y
array([12.495, 12.342, 12.149, 12.052, 12.133, 12.286, 12.395, 12.377, 12.15 , 11.746, 11.394, 11.259, 11.394, 11.674, 12.048, 12.118, 11.706, 11.103, 10.762, 10.914, 11.246, 11.458, 11.667, 11.801, 11.698, 11.346, 11.031, 10.894, 11.213, 11.437, 11.846, 12.064, 12.465, 12.374, 12.083, 11.801, 11.697, 11.775, 11.904, 11.976, 12.003, 11.897, 11.601, 11.367, 11.305, 11.381, 11.562, 11.791, 11.758, 11.483, 11.081, 10.809, 10.711, 10.985, 11.497, 11.895, 11.777, 11.584, 11.587, 11.75 , 11.637, 11.43 , 11.324, 10.862, 10.516, 10.642, 11.223, 11.698, 11.919, 12.168, 12.194, 12.117, 11.936, 11.796, 11.776, 11.741, 11.65 , 11.516, 11.182, 10.89 , 10.641, 10.495, 10.425, 10.37 , 10.227, 9.997, 9.82 , 9.953, 10.259, 10.425, 10.466, 10.229, 10.04 , 9.789, 9.904, 10.021, 10.129, 10.132, 10.211, 10.275, 10.3 , 10.373, 10.34 , 10.545, 10.711, 10.972, 11.378, 11.625, 11.632, 11.486, 11.395, 11.207, 11.049, 11.002, 10.925, 10.799, 10.65 , 10.468, 10.381, 10.307, 10.517, 10.849, 11.012, 10.949, 10.779, 10.83 , 10.868, 10.778, 10.651, 10.537, 10.504, 10.342, 9.949, 9.516, 9.085, 9.222, 9.711, 10.099, 10.129, 9.914, 9.692, 9.501]) - LONGITUDE(N_PROF)float64...
- long_name :
- Longitude of the station, best estimate
- standard_name :
- longitude
- units :
- degree_east
- valid_min :
- -180.0
- valid_max :
- 180.0
- axis :
- X
array([-26.079, -25.964, -25.929, -25.99 , -26.083, -26.26 , -26.416, -26.484, -26.603, -26.812, -27.142, -27.253, -27.32 , -27.273, -27.146, -26.947, -26.923, -26.967, -26.75 , -26.504, -26.465, -26.476, -26.407, -26.345, -26.283, -25.908, -25.435, -25.084, -24.841, -24.983, -25.227, -25.531, -25.723, -25.7 , -26.024, -26.234, -26.477, -26.618, -26.631, -26.664, -26.824, -26.956, -26.977, -26.951, -26.853, -26.688, -26.453, -26.027, -25.791, -25.797, -25.834, -25.969, -25.946, -25.825, -25.651, -25.551, -25.505, -25.159, -24.933, -24.799, -24.849, -24.996, -25.065, -25.117, -25.001, -24.769, -24.651, -24.645, -24.676, -24.693, -24.815, -25.06 , -25.509, -26.067, -26.707, -27.266, -27.77 , -28.32 , -28.696, -29.306, -29.889, -30.285, -30.461, -30.555, -30.609, -30.636, -30.758, -30.973, -31.272, -31.451, -31.593, -31.626, -31.748, -31.727, -31.621, -31.31 , -31.076, -31.027, -30.819, -30.785, -30.771, -30.717, -30.736, -30.764, -31.093, -31.423, -31.608, -31.317, -30.876, -30.706, -30.651, -30.689, -30.638, -30.634, -30.67 , -30.686, -30.73 , -30.761, -30.836, -30.926, -31.052, -31.091, -30.998, -30.727, -30.472, -30.32 , -30.276, -30.413, -30.68 , -31.025, -31.255, -31.382, -31.676, -31.602, -31.159, -30.777, -30.841, -30.82 , -30.688, -30.624, -30.499, -30.457]) - POSITION_QC(N_PROF)object...
- long_name :
- Quality on position (latitude and longitude)
- conventions :
- Argo reference table 2
array([b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1'], dtype=object) - POSITIONING_SYSTEM(N_PROF)object...
- long_name :
- Positioning system
array([b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS ', b'ARGOS '], dtype=object) - PROFILE_PRES_QC(N_PROF)object...
- long_name :
- Global quality flag of PRES profile
- conventions :
- Argo reference table 2a
array([b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'C', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'E', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A'], dtype=object) - PROFILE_TEMP_QC(N_PROF)object...
- long_name :
- Global quality flag of TEMP profile
- conventions :
- Argo reference table 2a
array([b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'C', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A'], dtype=object) - PROFILE_PSAL_QC(N_PROF)object...
- long_name :
- Global quality flag of PSAL profile
- conventions :
- Argo reference table 2a
array([b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'C', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'F', b'F', b'F', b'F', b'F', b'A', b'A', b'A', b'F', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'F', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A'], dtype=object) - VERTICAL_SAMPLING_SCHEME(N_PROF)object...
- long_name :
- Vertical sampling scheme
- conventions :
- Argo reference table 16
array([b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete [] '], dtype=object) - CONFIG_MISSION_NUMBER(N_PROF)float64...
- long_name :
- Unique number denoting the missions performed by the float
- conventions :
- 1...N, 1 : first complete mission
array([1., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2., 2.]) - PRES(N_PROF, N_LEVELS)float325.5 9.6 20.2 ... 1.948e+03 2e+03
- long_name :
- Sea water pressure, equals 0 at sea-level
- standard_name :
- sea_water_pressure
- units :
- decibar
- valid_min :
- 0.0
- valid_max :
- 12000.0
- C_format :
- %7.1f
- FORTRAN_format :
- F7.1
- resolution :
- 1.0
- axis :
- Z
array([[ 5.5, 9.6, 20.2, ..., 1900.4, 1950.2, 1998.4], [ 6.1, 10.1, 19.6, ..., 1900.1, 1949.8, 1997.8], [ 5.6, 9.8, 20.3, ..., 1899.3, 1949. , nan], ..., [ 6.3, 10. , 20.7, ..., 1900. , 1950.7, nan], [ 5.9, 8.8, 20.3, ..., 1897.7, 1950.2, nan], [ 5.7, 10.7, 20.7, ..., 1897.8, 1947.7, 1999.8]], dtype=float32) - PRES_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
array([[b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', nan], ..., [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', b'1']], dtype=object) - PRES_ADJUSTED(N_PROF, N_LEVELS)float325.3 9.4 20.0 ... 1.948e+03 2e+03
- long_name :
- Sea water pressure, equals 0 at sea-level
- standard_name :
- sea_water_pressure
- units :
- decibar
- valid_min :
- 0.0
- valid_max :
- 12000.0
- C_format :
- %7.1f
- FORTRAN_format :
- F7.1
- resolution :
- 1.0
- axis :
- Z
array([[ 5.3, 9.4, 20. , ..., 1900.2, 1950. , 1998.2], [ 6. , 10. , 19.5, ..., 1900. , 1949.7, 1997.7], [ 5.5, 9.7, 20.2, ..., 1899.2, 1948.9, nan], ..., [ 6.5, 10.2, 20.9, ..., 1900.2, 1950.9, nan], [ 6.1, 9. , 20.5, ..., 1897.9, 1950.4, nan], [ 5.9, 10.9, 20.9, ..., 1898. , 1947.9, 2000. ]], dtype=float32) - PRES_ADJUSTED_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
array([[b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', nan], ..., [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', b'1']], dtype=object) - PRES_ADJUSTED_ERROR(N_PROF, N_LEVELS)float32...
- long_name :
- Contains the error on the adjusted values as determined by the delayed mode QC process
- units :
- decibar
- C_format :
- %7.1f
- FORTRAN_format :
- F7.1
- resolution :
- 1.0
array([[2.4, 2.4, 2.4, ..., 2.4, 2.4, 2.4], [2.4, 2.4, 2.4, ..., 2.4, 2.4, 2.4], [2.4, 2.4, 2.4, ..., 2.4, 2.4, nan], ..., [2.4, 2.4, 2.4, ..., 2.4, 2.4, nan], [2.4, 2.4, 2.4, ..., 2.4, 2.4, nan], [2.4, 2.4, 2.4, ..., 2.4, 2.4, 2.4]], dtype=float32) - TEMP(N_PROF, N_LEVELS)float3226.94 26.94 26.93 ... 3.645 3.546
- long_name :
- Sea temperature in-situ ITS-90 scale
- standard_name :
- sea_water_temperature
- units :
- degree_Celsius
- valid_min :
- -2.5
- valid_max :
- 40.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
array([[26.939, 26.938, 26.934, ..., 3.772, 3.715, 3.64 ], [26.336, 26.336, 26.335, ..., 3.733, 3.682, 3.618], [25.494, 25.486, 25.481, ..., 3.707, 3.653, nan], ..., [28.814, 28.783, 28.726, ..., 3.693, 3.641, nan], [28.359, 28.355, 28.353, ..., 3.702, 3.625, nan], [27.837, 27.837, 25.937, ..., 3.693, 3.645, 3.546]], dtype=float32) - TEMP_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
array([[b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', nan], ..., [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', b'1']], dtype=object) - TEMP_ADJUSTED(N_PROF, N_LEVELS)float3226.94 26.94 26.93 ... 3.645 3.546
- long_name :
- Sea temperature in-situ ITS-90 scale
- standard_name :
- sea_water_temperature
- units :
- degree_Celsius
- valid_min :
- -2.5
- valid_max :
- 40.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
array([[26.939, 26.938, 26.934, ..., 3.772, 3.715, 3.64 ], [26.336, 26.336, 26.335, ..., 3.733, 3.682, 3.618], [25.494, 25.486, 25.481, ..., 3.707, 3.653, nan], ..., [28.814, 28.783, 28.726, ..., 3.693, 3.641, nan], [28.359, 28.355, 28.353, ..., 3.702, 3.625, nan], [27.837, 27.837, 25.937, ..., 3.693, 3.645, 3.546]], dtype=float32) - TEMP_ADJUSTED_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
array([[b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', nan], ..., [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', b'1']], dtype=object) - TEMP_ADJUSTED_ERROR(N_PROF, N_LEVELS)float32...
- long_name :
- Contains the error on the adjusted values as determined by the delayed mode QC process
- units :
- degree_Celsius
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
array([[0.0025, 0.0025, 0.0025, ..., 0.0025, 0.0025, 0.0025], [0.0025, 0.0025, 0.0025, ..., 0.0025, 0.0025, 0.0025], [0.0025, 0.0025, 0.0025, ..., 0.0025, 0.0025, nan], ..., [0.0025, 0.0025, 0.0025, ..., 0.0025, 0.0025, nan], [0.0025, 0.0025, 0.0025, ..., 0.0025, 0.0025, nan], [0.0025, 0.0025, 0.0025, ..., 0.0025, 0.0025, 0.0025]], dtype=float32) - PSAL(N_PROF, N_LEVELS)float3235.71 35.71 35.71 ... 34.96 34.96
- long_name :
- Practical salinity
- standard_name :
- sea_water_salinity
- units :
- psu
- valid_min :
- 2.0
- valid_max :
- 41.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
array([[35.71 , 35.709, 35.71 , ..., 34.977, 34.975, 34.973], [35.765, 35.765, 35.765, ..., 34.973, 34.972, 34.971], [35.836, 35.836, 35.836, ..., 34.972, 34.971, nan], ..., [35.086, 35.092, 35.105, ..., 34.963, 34.961, nan], [35.26 , 35.272, 35.285, ..., 34.963, 34.961, nan], [35.118, 35.12 , 35.776, ..., 34.964, 34.962, 34.96 ]], dtype=float32) - PSAL_QC(N_PROF, N_LEVELS)objectb'1' b'1' b'1' ... b'1' b'1' b'1'
- long_name :
- quality flag
- conventions :
- Argo reference table 2
array([[b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', b'1'], [b'1', b'1', b'1', ..., b'1', b'1', nan], ..., [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', nan], [b'1', b'1', b'1', ..., b'1', b'1', b'1']], dtype=object) - PSAL_ADJUSTED(N_PROF, N_LEVELS)float3235.7 35.7 35.7 ... 34.96 34.96
- long_name :
- Practical salinity
- standard_name :
- sea_water_salinity
- units :
- psu
- valid_min :
- 2.0
- valid_max :
- 41.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
array([[35.704 , 35.703 , 35.704 , ..., 34.971 , 34.969 , 34.967 ], [35.759 , 35.759 , 35.759 , ..., 34.967 , 34.966 , 34.965 ], [35.831 , 35.831 , 35.831 , ..., 34.967 , 34.966 , nan], ..., [35.101 , 35.108 , 35.127 , ..., 34.977 , 34.975 , nan], [35.275 , 35.287 , 35.304 , ..., 34.977 , 34.975 , nan], [35.11832, 35.13407, 35.77779, ..., 34.96395, 34.962 , 34.9599 ]], dtype=float32) - PSAL_ADJUSTED_QC(N_PROF, N_LEVELS)objectb'2' b'2' b'2' ... b'1' b'1' b'1'
- long_name :
- quality flag
- conventions :
- Argo reference table 2
array([[b'2', b'2', b'2', ..., b'2', b'2', b'2'], [b'2', b'2', b'2', ..., b'2', b'2', b'2'], [b'2', b'2', b'2', ..., b'2', b'2', nan], ..., [b'2', b'2', b'2', ..., b'2', b'2', nan], [b'2', b'2', b'2', ..., b'2', b'2', nan], [b'1', b'1', b'1', ..., b'1', b'1', b'1']], dtype=object) - PSAL_ADJUSTED_ERROR(N_PROF, N_LEVELS)float32...
- long_name :
- Contains the error on the adjusted values as determined by the delayed mode QC process
- units :
- psu
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
array([[0.01 , 0.01 , 0.01 , ..., 0.01 , 0.01 , 0.01 ], [0.01 , 0.01 , 0.01 , ..., 0.01 , 0.01 , 0.01 ], [0.01 , 0.01 , 0.01 , ..., 0.01 , 0.01 , nan], ..., [0.01 , 0.01 , 0.01 , ..., 0.01 , 0.01 , nan], [0.01 , 0.01 , 0.01 , ..., 0.01 , 0.01 , nan], [0.01 , 0.01445, 0.01 , ..., 0.01 , 0.01 , 0.01 ]], dtype=float32) - PARAMETER(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- List of parameters with calibration information
- conventions :
- Argo reference table 3
array([[[b'PRES ', b'TEMP ', b'PSAL ']], [[b'PRES ', b'TEMP ', b'PSAL ']], ..., [[b'PRES ', b'TEMP ', b'PSAL ']], [[b'PRES ', b'TEMP ', b'PSAL ']]], dtype=object) - SCIENTIFIC_CALIB_EQUATION(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- Calibration equation for this parameter
array([[[b'PRES_ADJUSTED(cycle i)=PRES (cycle i)-Surface Pressure(cycle i+1). ', b'TEMP_ADJUSTED = TEMP ', b'PSAL re-calculated using PRES_ADJUSTED.PSAL Cell Thermal Mass celltm_sbe41.PSAL_ADJUSTED=PSAL+DeltaS, DeltaS is calculated a potential conductivity (ref to 0 dbar) multiplicative adjustment term r. ']], [[b'PRES_ADJUSTED(cycle i)=PRES (cycle i)-Surface Pressure(cycle i+1). ', b' ', b'PSAL re-calculated using PRES_ADJUSTED.PSAL Cell Thermal Mass celltm_sbe41.PSAL_ADJUSTED=PSAL+DeltaS, DeltaS is calculated a potential conductivity (ref to 0 dbar) multiplicative adjustment term r. ']], ..., [[b'PRES_ADJUSTED(cycle i)=PRES (cycle i)-Surface Pressure(cycle i+1). ', b' ', b'PSAL re-calculated using PRES_ADJUSTED.PSAL Cell Thermal Mass celltm_sbe41.PSAL_ADJUSTED=PSAL+DeltaS, DeltaS is calculated a potential conductivity (ref to 0 dbar) multiplicative adjustment term r. ']], [[b'PRES_ADJUSTED(cycle i)=PRES (cycle i)-Surface Pressure(cycle i+1). ', b' ', b'PSAL re-calculated using PRES_ADJUSTED.PSAL Cell Thermal Mass celltm_sbe41. ']]], dtype=object) - SCIENTIFIC_CALIB_COEFFICIENT(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- Calibration coefficients for this equation
array([[[b'Surface pressure=0 dbar dbar ', b'Not applicable ', b'r = 0.99983997 (+/- 0.00007583), vertically averaged DeltaS = -0.0063 (+/- 0.0030) ']], [[b'Surface pressure=0 dbar ', b' ', b'r = 0.99984934 (+/- 0.00007133), vertically averaged DeltaS = -0.0059 (+/- 0.0028) ']], ..., [[b'Surface pressure=0 dbar ', b' ', b'r = 1.00036551 (+/- 0.00007129), vertically averaged DeltaS = 0.0143 (+/- 0.0028) ']], [[b'Surface pressure=0 dbar ', b' ', b'alpha = 0.0267, tau = 18.6 and Ascend velocity = 0.09. ']]], dtype=object) - SCIENTIFIC_CALIB_COMMENT(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- Comment applying to this parameter calibration
array([[[b'APEX float that not truncated negative pressure drift. Pressure adjusted by using pressure offset at the sea surface. Calibration error is manufacturer specified accuracy in dbar. ', b'No adjustment performed (values duplicated) ', b'Salinity recomputed for pressure offset.ed)Salinity corrected for CTM.Drift detected,adjusted salinity using WJO(2003), WOD2001 as database, mapping scales 8/4,4/2,sliding calibration window +/-20prof. ']], [[b'APEX float that not truncated negative pressure drift. Pressure adjusted by using pressure offset at the sea surface. Calibration error is manufacturer specified accuracy in dbar. ', b'none ', b'Salinity recomputed for pressure offset.Salinity corrected for CTM.Drift detected,adjusted salinity using WJO(2003), WOD2001 as database, mapping scales 8/4,4/2,sliding calibration window +/-20prof. ']], ..., [[b'APEX float that not truncated negative pressure drift. Pressure adjusted by using pressure offset at the sea surface. Calibration error is manufacturer specified accuracy in dbar. ', b'none ', b'Salinity recomputed for pressure offset.Salinity corrected for CTM.Drift detected,adjusted salinity using WJO(2003), WOD2001 as database, mapping scales 8/4,4/2,sliding calibration window +/-20prof. ']], [[b'APEX float that not truncated negative pressure drift. Pressure adjusted by using pressure offset at the sea surface. Calibration error is manufacturer specified accuracy in dbar. ', b'none ', b'Salinity recomputed for pressure offset.Salinity corrected for CTM. ']]], dtype=object) - SCIENTIFIC_CALIB_DATE(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- Date of calibration
- conventions :
- YYYYMMDDHHMISS
array([[[b'20161219132250', b'20161104203241', b'20170315130512']], [[b'20161219132250', b'20170315131707', b'20170315130512']], ..., [[b'20161219132250', b'20170315131949', b'20170315130513']], [[b'20161219132250', b'20170315131950', b'20161219132320']]], dtype=object) - HISTORY_INSTITUTION(N_HISTORY, N_PROF)object...
- long_name :
- Institution which performed action
- conventions :
- Argo reference table 4
array([], shape=(0, 142), dtype=object)
- HISTORY_STEP(N_HISTORY, N_PROF)object...
- long_name :
- Step in data processing
- conventions :
- Argo reference table 12
array([], shape=(0, 142), dtype=object)
- HISTORY_SOFTWARE(N_HISTORY, N_PROF)object...
- long_name :
- Name of software which performed action
- conventions :
- Institution dependent
array([], shape=(0, 142), dtype=object)
- HISTORY_SOFTWARE_RELEASE(N_HISTORY, N_PROF)object...
- long_name :
- Version/release of software which performed action
- conventions :
- Institution dependent
array([], shape=(0, 142), dtype=object)
- HISTORY_REFERENCE(N_HISTORY, N_PROF)object...
- long_name :
- Reference of database
- conventions :
- Institution dependent
array([], shape=(0, 142), dtype=object)
- HISTORY_DATE(N_HISTORY, N_PROF)object...
- long_name :
- Date the history record was created
- conventions :
- YYYYMMDDHHMISS
array([], shape=(0, 142), dtype=object)
- HISTORY_ACTION(N_HISTORY, N_PROF)object...
- long_name :
- Action performed on data
- conventions :
- Argo reference table 7
array([], shape=(0, 142), dtype=object)
- HISTORY_PARAMETER(N_HISTORY, N_PROF)object...
- long_name :
- Station parameter action is performed on
- conventions :
- Argo reference table 3
array([], shape=(0, 142), dtype=object)
- HISTORY_START_PRES(N_HISTORY, N_PROF)float32...
- long_name :
- Start pressure action applied on
- units :
- decibar
array([], shape=(0, 142), dtype=float32)
- HISTORY_STOP_PRES(N_HISTORY, N_PROF)float32...
- long_name :
- Stop pressure action applied on
- units :
- decibar
array([], shape=(0, 142), dtype=float32)
- HISTORY_PREVIOUS_VALUE(N_HISTORY, N_PROF)float32...
- long_name :
- Parameter/Flag previous value before action
array([], shape=(0, 142), dtype=float32)
- HISTORY_QCTEST(N_HISTORY, N_PROF)object...
- long_name :
- Documentation of tests performed, tests failed (in hex form)
- conventions :
- Write tests performed when ACTION=QCP$; tests failed when ACTION=QCF$
array([], shape=(0, 142), dtype=object)
- title :
- Argo float vertical profile
- institution :
- FR GDAC
- source :
- Argo float
- history :
- 2018-11-20T18:47:39Z creation
- references :
- http://www.argodatamgt.org/Documentation
- user_manual_version :
- 3.1
- Conventions :
- Argo-3.1 CF-1.6
- featureType :
- trajectoryProfile
